SPECIAL FEATURE
Computing Research that Changed the World: Reflections and Perspectives
March 25, 2009 | 8:45 am - 5:00 pm | Members' Room, Thomas Jefferson Building, Library of Congress
Zooming In On Life
GENE MYERS - HHMI
![]() Slides - 3.5 MB
Slides - 3.5 MB
![]() Download - 267 MB
Download - 267 MB
![]() Watch the Talk (18:09)
Watch the Talk (18:09)
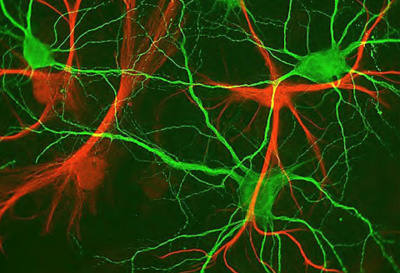 Biology has become an "information science," and computer science is transforming our understanding of biology.
Biology has become an "information science," and computer science is transforming our understanding of biology.
An intriguing challenge for computational biology is to understand how various components of a cell work, and how these cells are mapped in an organ such as the brain. The latter can be understood by developing novel imaging and microscopy technology, combined with clever data analysis. One technique that facilitates imaging is the mapping of the genome, which helps researchers to modify the genome to add fluorescent markers. The markers can then be used for tracking the protein products of these genes. This technique is being used to map the process of cell division in bacteria.
At Celera Genomics, the whole genome shotgun technique (where an entire genome is mapped at once using combinatorial inference methods) was developed over a period of years. Shotgun sequencing helped Celera map the human genome at a speed much faster than the Human Genome Project. The Celera approach replaced a large amount of physical work and instrumentation by computation. Computer science played a critical enabling role. Both hardware advances and algorithmic advances were essential. The genome project has produced many widely-used software tools, for instance, the BLAST software.
Looking forward, current hardware technology is far superior to that available when Celera started on this project: today, one can purchase single computers, each with processing power comparable to the entire power available to Celera less than a decade ago. In current sequencing efforts, the bottleneck is often not the hardware, but the algorithms and software. The existing software and algorithms often strain the available resources, but there is a shortage of experts in informatics and software to fix this problem.
The current focus of research in sequencing is to personalize the genome. The goal is to determine "your genome for $1000." This achievement will enable the discovery of novel genotype-phenotype correlations, i.e., the correlation between genetic properties and observable properties such as blood pressure. This is a difficult problem, but its solution will advance personalized medicine by tailoring drugs and treatments to specific genotype-phenotype combinations. (It is not certain that this vision of personalized medicine will pan out, since human beings are complex beings, and the phenotype is often determined by factors which are hard to understand and control.)
A related research focus is to sequence as many organisms as possible, which will aid in the genetic study of evolution. Progress on this topic is critical for several reasons: cancer is a form of evolution, and so is pathogenesis, that is, the evolution of pathogens to adapt to treatments and other chemicals. Better understanding of these phenomena might lead to better treatments.
A final focus is synthetic biology, which is the creation of new sequences and new bacteria with useful properties. Synthetic biology has enormous potential, and also leads to several thorny ethical and policy questions.
At a larger scale, another research challenge is to make an atlas of different organs by mapping where cells are located. There is increasing research interest in the brain. A starting point is to map the neurons in the fly and mouse brains and to build a complete model. Just as in sequencing, the goal is to find structure and not function: the hope is that the structure itself will lend itself to useful applications.
Genomics has become the key to molecular biology. The ability to do recombinant genetics on the whole genome enables biologists to manipulate genes and control expression. Computation has become the key to genomics, making biology a big data science.
Computation has become the bottleneck to further progress in biology. Bio-imaging informatics, visualizing the data and the processes, is a major challenge. Dedicated machines are needed for this imaging. Cloud computing is not an alternative, since the bottleneck is often to get the data to the cloud. Furthermore, the amount of data is too massive to be archived. The goal has therefore become to extract key features from the data, and throw the raw data away. This data collection and imaging effort also leads to understanding biophysics better, for instance, the process of cell division. The challenges for computing research are abundant and the payoff is potentially enormous.

